This page is part of the HLA Reporting Implementation Guide (v0.0.1: Trial Use: 1) based on FHIR R4. This is the current published version. For a full list of available versions, see the Directory of published versions 
Home
Contents:
Human leukocyte antigen (HLA) genotyping is fundamental for research and clinical practice in immunogenetics and histocompatibility. This HLA Reporting Implementation Guide (IG) describes how to report HLA genotyping results using HL7 FHIR. It relies heavily on the Genomics Reporting IG developed by the HL7 Clinical Genomics Workgroup, constraining profiles for GeneticsReport, Genotype and Haplotype. It also constrains the MolecularSequence resource to allow reporting sequencing results together with novel variants.
Profiles in this IG
Building a report
A complete HLA genotyping report is typically contained in a FHIR transaction Bundle containing individual resources conforming to the profiles described in this IG. Examples of full transaction bundles included below. Minimally, it would contain an HLA Summary Report and one or more HLA Genotype observations.
Summary Report and Genotype Observation
In a simple case, the HLA Summary Report refers to an HLA Genotype observation for each HLA gene. The HLA alleles found in the genotype are assigned to alleles found in the the Immuno Polymorphism Database (IPD) through the international ImMunoGeneTics (IMGT) project (found here) and expressed as a Genotype List String (GLString) using a GLString Code.
If it is necessary to report the molecular sequence that was used to derive the HLA Genotype observation, one or more HLA Sequence profiles can be used. This is shown in Fig 1.
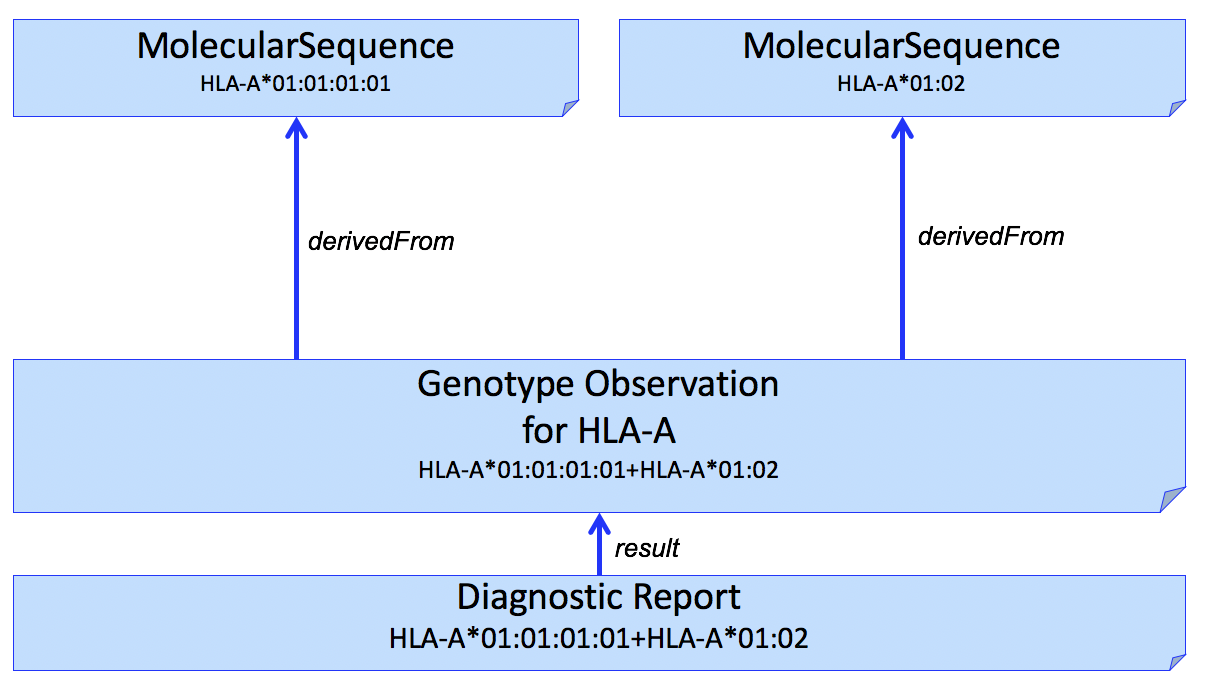
Figure 1
HLA - Adding the individual alleles
It is often desirable to identify sequence data that was used to assert or assign individual HLA alleles. This can be done by adding another Observation to capture this, shown in Figure 2.
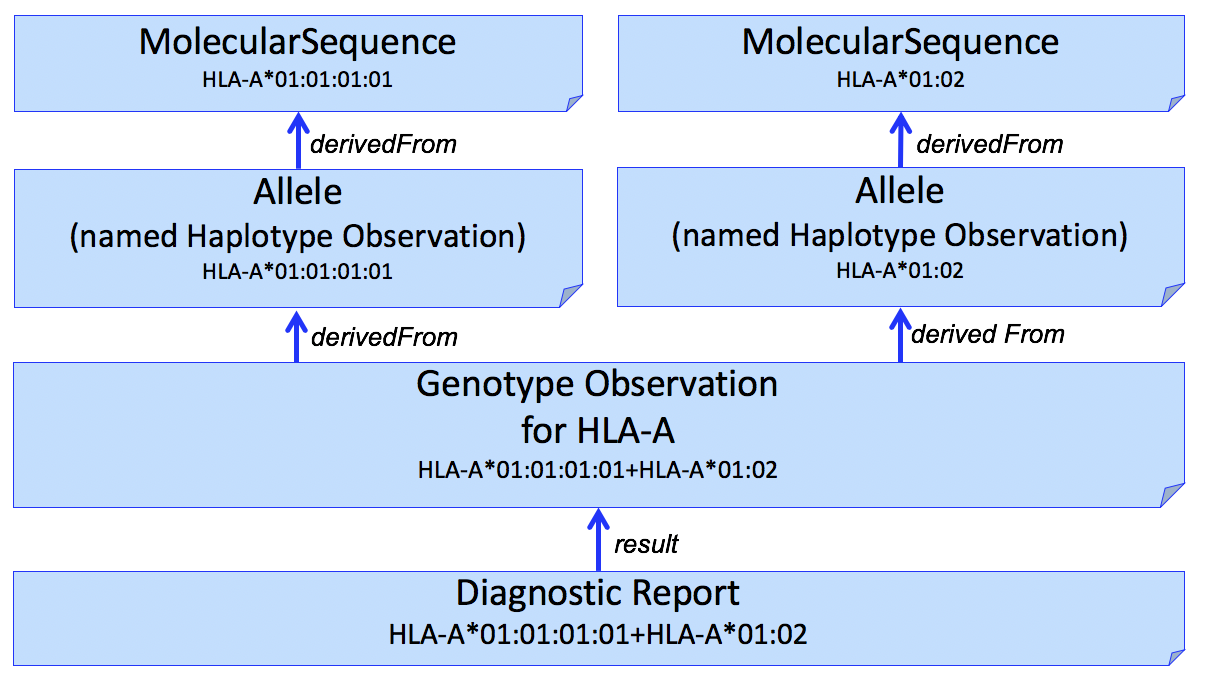
Figure 2
The example in this figure is a simplistic and the actual results often includes multiple sequences (e.g., exons 2 and 3). Allele assignment is a complex process and may result in ambiguous results (see Reporting Ambiguity below).
We use the HLA Allele profile for Allele assignment. This profile is derived from the Haplotype profile. In domains other than HLA (e.g. Pharmacogenomics), it is common to consider gene-level alleles as named haplotypes of a set of smaller variants (Variation Modelling Collaboration Data Model and Specification). This is not how the HLA community considers Haplotype, which is typically used to describe a set of gene-level alleles from different HLA genes that are found on the same DNA molecule. To be able to represent this latter description, this IG allows the representation "haplotypes of haplotypes." (example not shown)
HLA - reporting multiple HLA genes
This pattern can be extended to multiple HLA genes in a single report. An example for HLA-A, HLA-B, and HLA-C is shown in Figure 3.
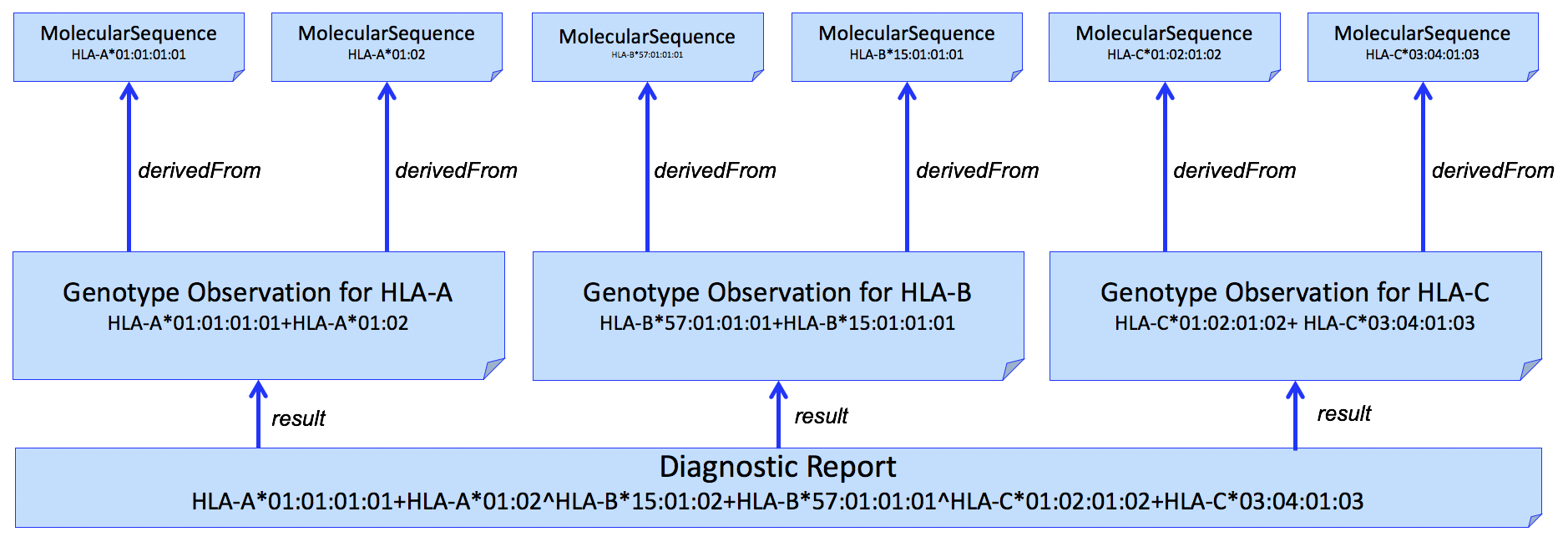
Figure 3
And adding individual alleles...
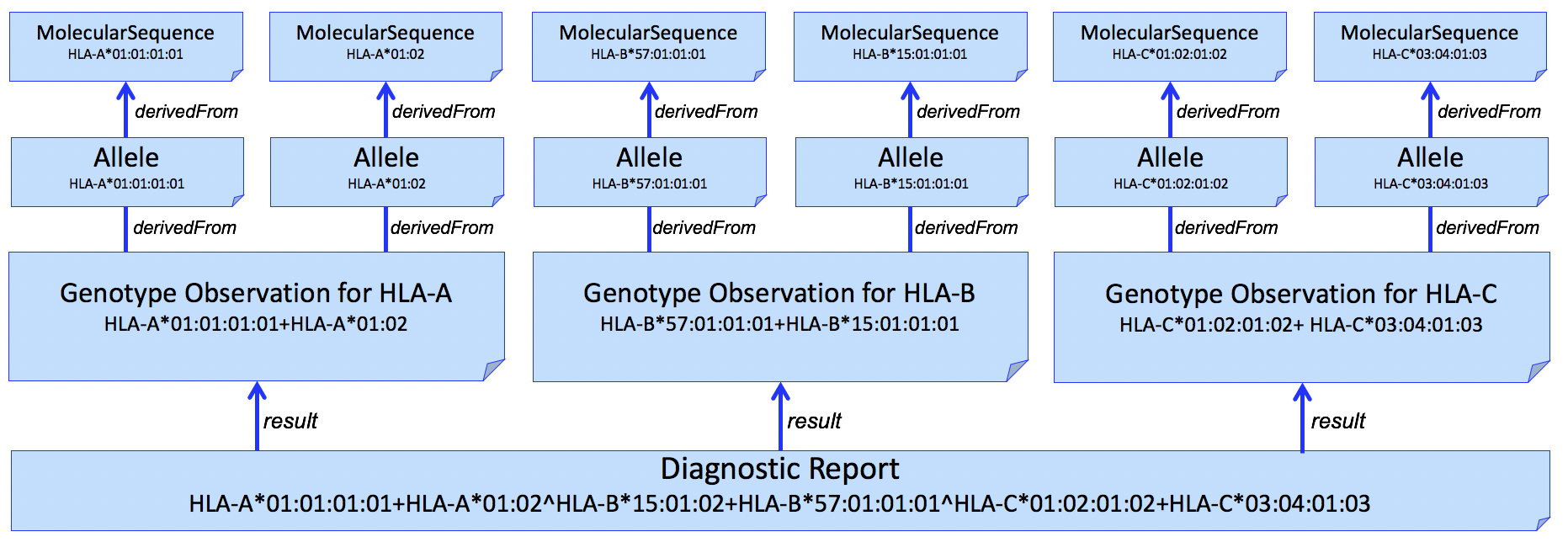
Figure 4
Building a bundle
Once individual resources are created, including references to each other, a FHIR transaction bundle may be created. A skeleton of such a FHIR bundle containing a Summary Report and a Genotype observation is shown below
<Bundle>
<type value="transaction"/>
<!-- each entry is an action to take in the transaction -->
<!-- HLA Summary Report -->
<entry>
<fullUrl value="urn:uuid:[unique uuid for HLA Summary]"/>
<resource>
<DiagnosticReport>
<meta>
<profile
value="http://fhir.nmdp.org/ig/hla-reporting/StructureDefinition/hla-summary-report"
/>
</meta>
...snip...
<result>
<reference value="urn:uuid:[unique uuid for HLA Genotype found in this bundle]"/>
</result>
</DiagnosticReport>
</resource>
<request>
<method value="POST"/>
<url value="DiagnosticReport"/>
</request>
</entry>
<!-- HLA Genotype Observation -->
<entry>
<fullUrl value="urn:uuid:[unique uuid for HLA Genotype]"/>
<resource>
<Observation>
<meta>
<profile
value="http://fhir.nmdp.org/ig/hla-reporting/StructureDefinition/hla-genotype"
/>
</meta>
...snip...
<derivedFrom>
<reference value="urn:uuid:[unique uuid for HLA Allele found in this bundle]"/>
</derivedFrom>
...etc...
</Observation>
</resource>
<request>
<method value="POST"/>
<url value="Observation"/>
</request>
</entry>
...other entries...
</Bundle>
HLA - Reporting Ambiguity
HLA allele assignment is often ambiguous. Genotype ambiguity results from an inability to establish chromosomal phase between identified polymorphisms. Allele ambiguity results when the polymorphisms that distinguish alleles fall outside of the regions assessed by the genotyping system.
Allele ambiguity is often captured using NMDP Multiple Allele Codes. Because of limitations of this system, including introducing further ambiguity into the report, the GL String is preferred. This format uses a hierarchical set of operators to describe the relationships between alleles, lists of possible alleles, phased alleles, genotypes, lists of possible genotypes, and multilocus unphased genotypes, without losing typing information or increasing typing ambiguity. To use this format, the GL String Code system can be used, which embeds this GL String into a format containing the gene system, and version of the nomenclature used within the GL String.
For example, this valueCodeableConcept would be used to describe a genotype for HLA-B, based on the 3.31.0 release of the IMGT-IPD/HLA database, that is ambiguous for one of the alleles in the genotype (HLA-B*07:02:01:01/HLA-B*07:02:01:03), and unambiguous for the other (HLA-B*13:02:01:01).
<valueCodeableConcept>
<coding>
<system value="http://glstring.org"/>
<code value="hla#3.31.0#HLA-B*07:02:01:01/HLA-B*07:02:01:03+HLA-B*13:02:01:01"/>
</coding>
</valueCodeableConcept>
HLA - LOINC and additional codings
The Observations used in this IG require specific codes. For example, the HLA Genotype observation requires a code.coding using a LOINC code that reprepresents "Genotype display name". To indicate that this genotype is for HLA-A, we use the component for Gene Studied. Together this code.coding and the component indicate that this Observation is a Genotype for HLA-A. This 'post-coordination' of the Observation code allows for reporting for any genotype and gene name combination.
<code>
<coding>
<system value="http://loinc.org"/>
<code value="84413-4"/>
<display value="Genotype display name"/>
</coding>
</code>
...
<component>
<code>
<coding>
<system value="http://loinc.org"/>
<code value="48018-6"/>
<display value="Gene studied [ID]"/>
</coding>
</code>
<valueCodeableConcept>
<coding>
<system value="http://www.genenames.org/geneId"/>
<code value="HGNC:4931"/>
<display value="HLA-A"/>
</coding>
</valueCodeableConcept>
</component>
Often a lab will use another LOINC code to represent this, or use a local code system. This can be included as an additional coding. For example, at the time of this writing, there are ten different LOINC codes related to typing of HLA-A. As long as the alternative coding is conceptual the same or is subsumed by the other, an additional code.coding can be included if the lab or healthcare system uses it in their current system. In the following example, the LOINC code 13298-5 representing "HLA-A [Type]" is added as an additional code.coding that is subsumed by the more general "Genotype Display Name".
<code>
<coding>
<system value="http://loinc.org"/>
<code value="84413-4"/>
<display value="Genotype display name"/>
</coding>
<coding>
<system value="http://loinc.org"/>
<code value="13298-5"/>
<display value="HLA-A [Type]"/>
</coding>
</code>
...
<component>
<code>
<coding>
<system value="http://loinc.org"/>
<code value="48018-6"/>
<display value="Gene studied [ID]"/>
</coding>
</code>
<valueCodeableConcept>
<coding>
<system value="http://www.genenames.org/geneId"/>
<code value="HGNC:4931"/>
<display value="HLA-A"/>
</coding>
</valueCodeableConcept>
</component>
HLA - Examples
- HLA-A,B Summary Report
- Genotype Observation for HLA-A
- HLA-A Allele Observation for HLA-A*01:01:01:01
- HLA-A Allele Observation for HLA-A*01:02
- HLA-A MolecularSequence for HLA-A*01:01:01:01
- HLA-A MolecularSequence for HLA-A*01:02
- Genotype Observation for HLA-B
- HLA-A,B Transaction Bundle containing all of above